Deciphering the signature of seedborne fungi linked to rice sheath rot disease: insights from ITS rDNA sequencing analysis
Main Article Content
Abstract
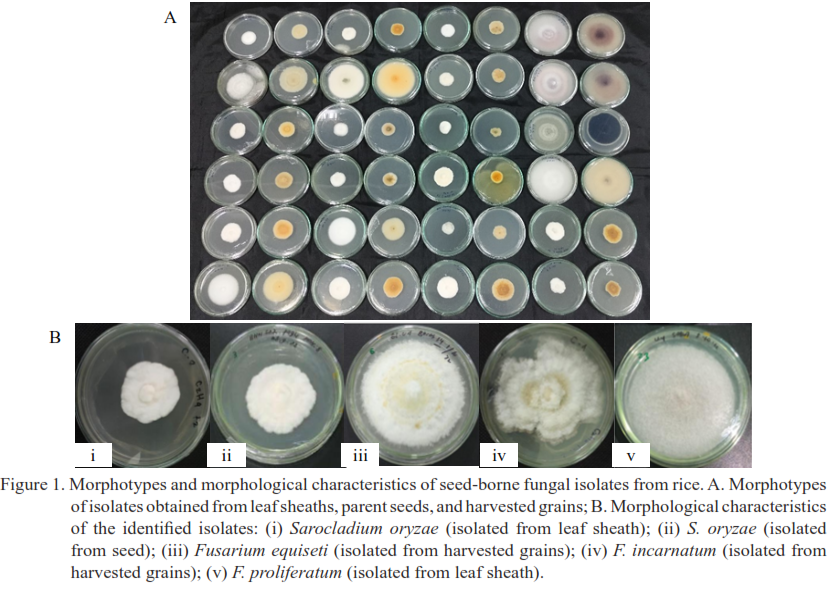
Sarocladium oryzae and Fusarium spp. are the causal agents of sheath rot, a re-emerging rice disease that has recently gained importance in Indonesia and can cause yield losses of up to 85%. Both pathogens are seedborne, making their accurate identification and management essential. Conventional morphological identification is time-consuming and often inaccurate due to overlapping symptoms among fungal species. In this study, we demonstrated the seedborne transmission of sheath rot pathogens and provided novel insights by highlighting the predominance of F. equiseti and the detection of infections in asymptomatic seeds. A total of 75 fungal isolates were obtained from rice leaf sheaths, seeds, and harvested grains across CMS, inbred, and hybrid rice varieties. ITS rDNA sequencing identified 42 isolates as S. oryzae and 33 as Fusarium spp., including F. equiseti (29), F. incarnatum (1), and F. proliferatum (3). The detection of these pathogens in both pre-planting seed samples and post-harvest grains demonstrates their ability to spread through seeds. Importantly, their presence in asymptomatic seeds and grains indicates that routine visual inspection is insufficient for seed health monitoring.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
References
Abbas HK, Cartwright RD, Shier WT, Abouzied MM, Bird CB, Rice LG, Ross PF, Sciumbato GL, & Meredith FI. 1998. Natural occurrence of fumonisins in rice with Fusarium sheath rot disease. Plant Dis. 82(1): 22–25. https://doi.org/10.1094/PDIS.1998.82.1.22
Abd-Elsalam KA, Aly IN, Abdel-Satar MA, Khalil MS, & Verreet JA. 2003. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr. J. Biotechnol. 2(4): 82–85. https://doi.org/10.5897/AJB2003.000-1016
Altschul SF, Gish W, Miller E, Meyers EW, & Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215(3): 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Aoki T, O’Donnell K, & Geiser DM. 2014. Systematics of key phytopathogenic Fusarium species: Current status and future challenges. J. Gen. Plant Pathol. 80: 189–201. https://doi.org/10.1007/s10327-014-0509-3
Bigirimana VdP, Hua GKH, Nyamangyoku OI, & Höfte M. 2015. Rice sheath rot: An emerging ubiquitous destructive disease complex. Front. Plant Sci. 6: 1066. https://doi.org/10.3389/fpls.2015.01066
Bruns TD, White TJ, & Taylor JW. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Sys. 22: 525–564. https://doi.org/10.1146/annurev.es.22.110191.002521
BSCI. 2020. Phylogenetic Trees. https://researchguides.library.vanderbilt.edu/bsci1511L. Accessed 22 September 2023.
Castellá G & Cabañes FJ. 2014. Phylogenetic diversity of Fusarium incarnatum-equiseti species complex isolated from Spanish wheat. Antonie Van Leeuwenhoek. 106: 309–317. https://doi.org/10.1007/s10482-014-0200-x
Chowdhury MTI, Mian MS, Mia MAT, Rafii MY, & Latif MA. 2015. Agro-ecological variations of sheath rot disease of rice caused by Sarocladium oryzae and DNA fingerprinting of the pathogen’s population structure. Genet. Mol. Res. 14(4): 18140–18152. https://doi.org/10.4238/2015.December.23.1
Desjardins AE, Manandhar HK, Plattner RD, Manandhar GG, Poling SM, & Maragos CM. 2000. Fusarium species from Nepalese rice and production of mycotoxins and gibberellic acid by selected species. Appl. Environ. Microbiol, 66(3): 1020–1025. https://doi.org/10.1128/AEM.66.3.1020-1025.2000
Dossou B & Silue D. 2017. Rice pathogens intercepted on seeds originating from 11 African countries and from the USA. SST. 46(1): 31–40. https://doi.org/10.15258/sst.2018.46.1.03
Fisher PJ & Petrini O. 1992. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.). New Phytol. 120(1): 137–143. https://doi.org/10.1111/j.1469-8137.1992.tb01066.x
Hall TA. 1999. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98.
Hibbett DS. 1992. Ribosomal RNA and fungal systematics. Trans. Mycol. Soc. Japan. 44(4): 533–556.
Kumar S, Milstein Y, Brami Y, Elbaum R, & Yermiyahu U. 2020. Silicon nanoparticles increase disease resistance and grain yield in rice. Sci Rep. 10: 1238. https://doi.org/10.1038/s41598-020-58253-4
Lee FN & Rush MC. 1983. Rice sheath blight: A major rice disease. Plant Dis. 67(7): 829–832. https://doi.org/10.1094/PD-67-829
Mancini V, Murolo S, & Romanazzi G. 2016. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 65(5): 691–703. https://doi.org/10.1111/ppa.12515
Marín P, Moretti A, Ritieni A, Jurado M, Vázquez C, & González-Jaén MT. 2012. Phylogenetic analyses and toxigenic profiles of Fusarium equiseti and Fusarium acuminatum isolated from cereals from Southern Europe. Food Microbiol. 31(2): 229–237. https://doi.org/10.1016/j.fm.2012.03.014
Mew TW & Gonzales P. 2002. A Handbook of Rice Seedborne Fungi. International Rice Research Institute, and Enfield NH (USA). Science Publishers. Los Banos.
Molla KA, Karmakar S, Molla J, Bajaj P, Varshney RK, Datta SK, & Datta K. 2020. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 18(4): 895–915. https://doi.org/10.1111/pbi.13312
Nair R. 1976. Incidence of sheath rot in rice-a potential problem for Sambalpur, Orissa. International Rice Research Newsletter. 1(1): 19.
Nayogyani W & Kasiamdari R. 2022. Characterization of the sheath blight complex of fungi in rice (Oryza sativa L.). Egypt. J. Bot. 62(3): 739–745. https://doi.org/10.21608/ejbo. 2022.115576.1878
Ou SH. 1985. Rice Diseases. 2nd Edition, Commonwealth Mycological Institute. Kew.
Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, et al. 2019. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95(1): 1–273. https://doi.org/10.1007/s13225-019-00421-w
Prabhukarthikeyan SR, Keerthana U, Nagendran K, Yadav MK, Parameswaran C, Panneerselvam P, & Rath PC. 2020. First report of Fusarium proliferatum causing sheath rot disease of rice in Eastern India. Plant Dis. 105(3): 704. https://doi.org/10.1094/PDIS-08-20-1846-PDN
Pramunadipta S, Widiastuti A, Wibowo A, Suga H, & Priyatmojo A. 2020. Short communication: Sarocladium oryzae associated with sheath rot disease of rice in Indonesia. Biodiversitas. 21(3): 1243–1249. https://doi.org/10.13057/biodiv/d210352
Pramunadipta S, Widiastuti A, Wibowo A, Suga H, & Priyatmojo A. 2022. Identification and pathogenicity of Fusarium spp. associated with the sheath rot disease of rice (Oryza sativa) in Indonesia. J. Plant Pathol. 104: 251–267. https://doi.org/10.1007/s42161-021-00988-x
Reddy OR & Sathyanarayana N. 2001. Seed-borne fungi of rice and quarantine significance. In: Sreenivasaprasad S & Johnson R (Eds.). Major Fungal Diseases of Rice. pp. 331–345. Springer. Dordrecht. https://doi.org/10.1007/978-94-017-2157-8_24
Reddy MM, Reddy CS, & Singh BG. 2000. Effect of sheath rot disease on qualitative characters of rice grain. J. Mycol. Pl. Pathol. 30(1): 68–72.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. PNAS. 109(16): 6241–6246. https://doi.org/10.1073/pnas.1117018109
Singh R & Vishunavat K. 2015. Seed transmission of Sarocladium oryzae and Fusarium moniliforme in different genotypes of rice. Int. J. Plant Prot. 8(2): 397–399. https://doi.org/10.15740/HAS/IJPP/8.2/397-399
Tamura K, Stecher G, & Kumar S. 2021. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7): 3022–3027. https://doi.org/10.1093/molbev/msab120
Tamura K, Nei M, & Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 101(30): 11030–11035. https://doi.org/0.1073/pnas.0404206101
Tiwari 2016. Seed health: importance, concepts, and diagnostics. In: Kumar P, Gupta VK, Tiwari AK, & Kamle M (Eds.). Current Trends in Plant Disease Diagnostics and Management Practices. pp. 207–219 Springer. Berlin. https://doi.org/10.1007/978-3-319-27312-9
Ward E, Kanyuka K, Motteram J, Kornyukhin D, & Adams MJ. 2005. The use of conventional and quantitative real-time PCR assays for Polymyxa graminis to examine host plant resistance, inoculum levels and intraspecific variation. New Phytol. 165(5): 875–885. https://doi.org/10.1111/j.1469-8137.2004.01291.x
White TJ, Bruns T, Lee S, & Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, & White TJ (Eds.). PCR Protocols: A Guide to Methods and Applications. pp. 315–322. Academic Press, Inc. United States. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Yao C, Frederiksen RA, & Magill CW. 1992. Length heterogeneity in ITS 2 and the methylation status of CCGG and GCGC sites in the rRNA genes of the genus Peronosclerospora. Curr. Genet. 22(5): 415–420. https://doi.org/ 10.1007/BF00352443