Molecular identification of oomycetes related to horticultural crops in Southern Sumatera and Java, Indonesia
Main Article Content
Abstract
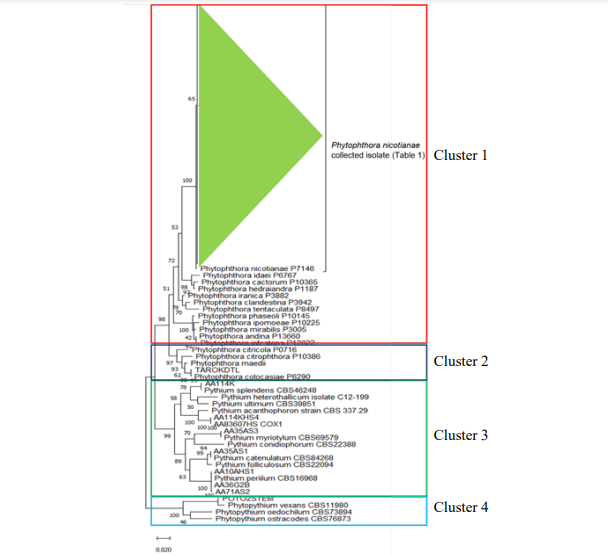
Indonesia is an agricultural country with more than 30 million farmers nationwide most of it with poor disease management. An identification of a pathogen is the first step to establish efficient management strategies for disease control. In this study, we survey the diversity of oomycetes in horticulture. Samples were collected from 19 sites around Lampung, Sumatera and Java Islands. The oomycetes were isolated from rhizosphere soils sample and from symptomatic plants tissues. One hundred and twelve isolates belonging to two Phytophthora spp., three Pythium spp., and one Phytophythium sp. were identified. Phytophthora nicotianae was a predominant species from pineapple but also found in cabbage, chilli, and chrysanthemum. P. colocasiae were isolated from taro in central java, Phytopythium vexans were isolated from potato in Central Java, while Pythium acanthophoron, Py. myriotylum, Py. splendens, and Py. catenulatum were isolated from soil in pineapple farms.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
References
Afandi A, Murayama E, Yin-Ling, Hieno A, & Kageyama K. 2018. Population structures of the water-borne plant pathogen Phytopythium helicoides reveal its possible origins and transmission modes in Japan. PLoS ONE. 13(12): e0209667. https://doi.org/10.1371/journal.pone.0209667
Agustina I, Pinem MI, & Zahara F. 2013. Uji efektivitas jamur antagonis Trichoderma sp. dan Gliocladium sp. untuk mengendalikan penyakit lanas (Phytophthora nicotianae) pada tanaman tembakau deli (Nicotiana tabaccum L.) [The test of the effectiviveness of Trichoderma sp. and Gliocladium sp. antagonism fungi is aimed to control lanas disease (Phytophthora nicotianae) in deli tobacco plants (Nicotiana tabaccum L.)]. Jurnal Online Agroekoteknologi. 1(4): 1130?1142.
Bala K, Robideau GP, Désaulniers N, de Cock AWAM, & Lévesque CA. 2010. Taxonomy, DNA barcoding and phylogeny of three new species of Pythium from Canada. Persoonia. 25: 22– 31. https://doi.org/10.3767/003158510X524754
Berlian I & Setyawan B. 2017. Gugur daun tanaman karet akibat penyakit Phytophthora spp.di perkebunan karet Jember, Jawa Timur [Incidence of leaf fall caused by Phytophthora spp. on rubber tree in Jember, East Java]. Warta Perkaretan. 36(2): 121–136. https://doi.org/10.22302/ppk.wp.v36i2.366
Blaha G, Hall G, Warokka JS, Concibido E, Ortiz-Garcia C. 1994. Phytophthora isolates from coconut plantations in Indonesia and Ivory Coast: characterization and identification by morphology and isozymes analysis. Mycol. Res. 98: 1379–1389. https://doi.org/10.1016/S0953-7562(09)81067-8
Derevnina L, Petre B, Kellner R, Dagdas YF, Sarowar MN, Giannakopoulu A, De la Concepcion JC, Chaparro-Garcia A, Pennington HG, van West P, & Kamoun S. 2016. Emerging oomycete threats to plants and animals. Phil. Trans R. Soc. B. 371(1709): 20150459. https://doi.org/10.1098/rstb.2015.0459
Gallup CA, Sullivan MJ, & Shew HD. 2006. Black Shank of Tobacco. The Plant Health Instructor. https://doi.org/10.1094/PHI-I-2006-0717-01
Goss EM, Tabima JF, Cooke DEL, Restrepo S, Fry WE, Forbes GA, Fieland VJ, Cardenas M, & Grünwald NJ. 2014. The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. PNAS. 111(24): 8791?8796. https://doi.org/10.1073/pnas.1401884111
Grünwald NJ, Garbelotto M, Goss EM, Heungens K, & Prospero S. 2012. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol. 20(3): 131?138. https://doi.org/10.1016/j.tim.2011.12.006
Kroon LPNM, Brouwer H, de Cock AWAM, & Govers F. 2012. The Genus Phytophthora Anno 2012. Phytopathology. 102(4): 348?364. https://doi.org/10.1094/PHYTO-01-11-0025
Lestari FW, Suharjono, & Arumingtyas EL. 2013. Phylogenetic identification of pathogenic fungi from apple in Batu City, Malang, Indonesia. Adv. Microbiol. 3(1): 69?75. https://doi.org/10.4236/aim.2013.31011
Lévesque CA & De Cock AWAM. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108(12): 1363–1383. https://doi.org/10.1017/S0953756204001431
Manohara D, Mulya K, Purwantara A, & Wahyuno D. 2004. Phytophthora capsici on Black Pepper in Indonesia. In: Drenth A & Guest DI (eds). Diversity and Management of Phytophthora in Southeast Asia. ACIAR Monograph 114. Australian Center for International Agricultural Research. Carnberra
Marpaung AE, Silalahi FE, & Purba EIY. 2010. Identifikasi patogen penyebab busuk pangkal batang pada tanaman jeruk di Tanah Karo [Identification of pathogens causing stem rot on citrus plants in Tanah Karo]. J. Hort. 20(3): 262–273.
Martin FN & Tooley PW. 2003. Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia. 95(2): 269–284. https://doi.org/10.1080/15572536.2004.11833112
Morita Y & Tojo M. 2007. Modifications of PARP medium using fluazinam, miconazole, and nystatin for detection of Pythium spp. in soil.Plant Disease : The American Phytopathological Society. 91(12): 1591–1599. https://doi.org/10.1094/PDIS-91-12-1591
Robideau GP, De Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Désaulniers N, Eggertson QA, Gachon CMM, Hu CH, Küpper FC, Rintoul TL, Sarhan E, Verstappen ECP, Zhang Y, Bonants PJM, Ristaino JB, & Lévesque CA. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Res. 11(6): 1002–1011. https://doi.org/10.1111/j.1755-0998.2011.03041.x
Santoso PJ, Aryantha INP, Pancoro A, & Suhandono S. 2015. Identification of Pythium and Phytophthora associated with durian (Durio sp.) in Indonesia: their molecular and morphological characteristics and distribution. Asian J. Plant Pathol. 9(2): 59–71. https://doi.org/10.3923/ajppaj.2015.59.71
Shew D & Gallup C. 2015. PROTOCOL 01-05.1: Isolation of Phytophthora nicotianae from soil. In: Ivors KL (Ed.). Laboratory Protocols for Phytophthora Species. pp. 1–3. APS PRESS. https://doi.org/10.1094/9780890544969.01.05.1
Tamura K, Stecher G, & Kumar S. 2021. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7): 3022–3027. https://doi.org/10.1093/molbev/msab120
Umayah A & Purwantara A. 2006. Identifikasi isolat Phytophthora asal kakao [Identification of isolates of Phytophthora from cocoa]. Menara Perkebunan. 74(2): 75–85. https://doi.org/10.22302/ppbbi.jur.mp.v74i2.108
Vanegtern B, Rogers M, & Nelson S. 2015. Black Pod Rot of Cacao Caused by Phytophthora palmivora. UH-CTHAR. PD–108. University of Hawaii. Hawaii
Vermeulen H & Bustamam M. 1977. Association of Pythium splendens with wilting and root rot of Chinese cabbage in Indonesia. Bulletin Penelitian Hortikultura. 13–17.
Villa NO, Kageyama K, Asano T, & Suga H. 2006. Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and ?-tubulin gene sequences. Mycologia. 98(3): 410–422. https://doi.org/10.1080/15572536.2006.11832676
Waterhouse GM & Waterston JM. 1966. Pythium splendens. [Descriptions of Fungi and Bacteria]. CABI International, Wallingford. https://doi.org/10.1079/DFB/20056400120
Yang X & Hong C. 2018. Differential usefulness of nine commonly used genetic markers for identifying Phytophthora species. Front. Microbiol. 03 October 2018. https://doi.org/10.3389/fmicb.2018.02334